Some self-explaining script examples can be found here.
Groovy code con be executed directly in Orbit via Tools -> Script Editor.
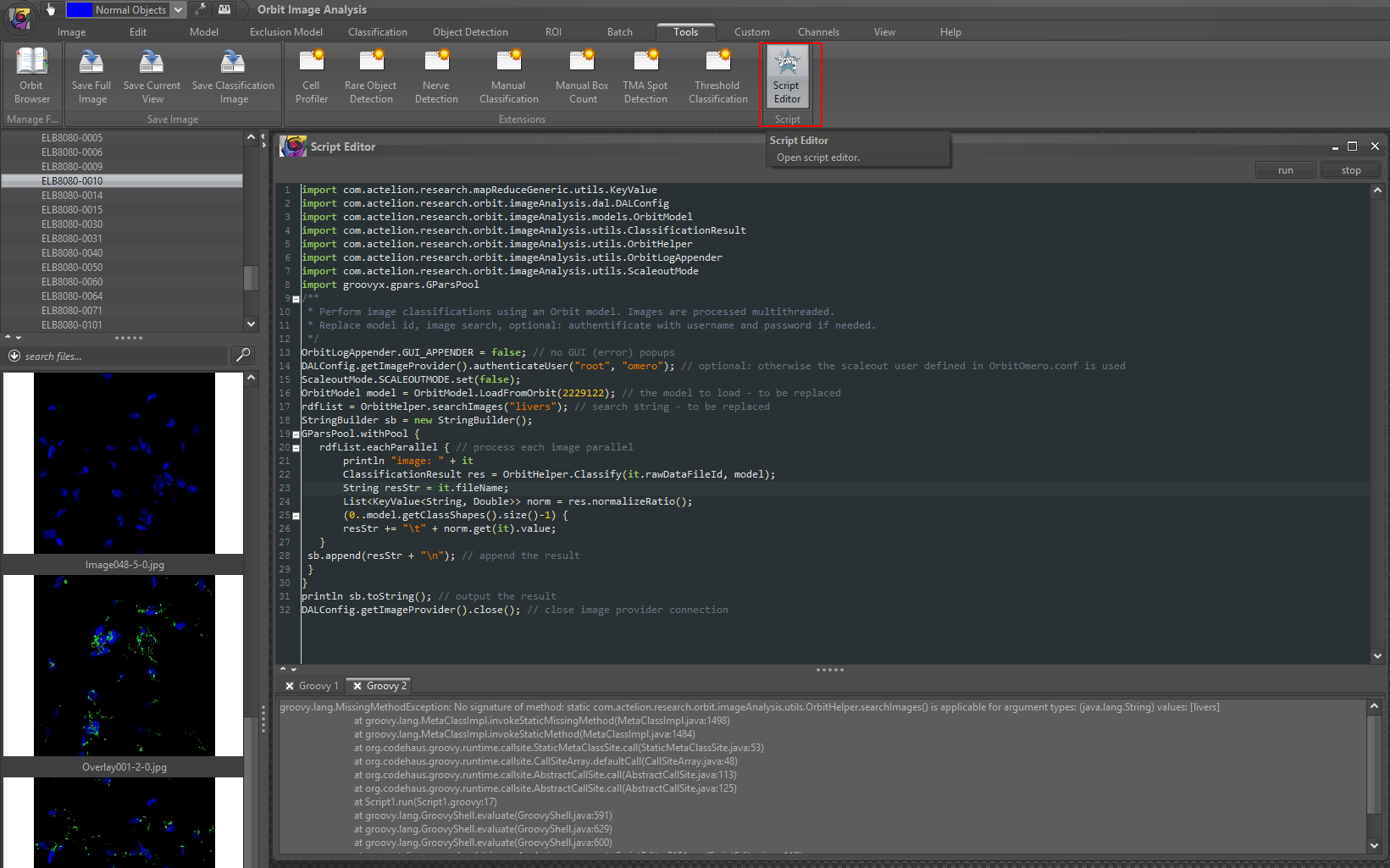
It runs within the current Java context and has the full Orbit classpath available.
The nice thing here is that OrbitImageAnalysis (the main controller class) is a singleton and can be accessed via
OrbitImageAnalysis.getInstance();
Thus, it is easy to access the current model or open image in the GUI:
import com.actelion.research.orbit.imageAnalysis.components.ImageFrame
import com.actelion.research.orbit.imageAnalysis.components.OrbitImageAnalysis
import com.actelion.research.orbit.imageAnalysis.models.OrbitModel
final OrbitImageAnalysis OIA = OrbitImageAnalysis.getInstance();
ImageFrame iFrame = OIA.getIFrame();
println("selected image: " + iFrame);
OrbitModel model = OIA.getModel();
println("active model: " + model);
This code can be provided easily to your users and they can execute it themselves.
In addition you can use the Orbit command line Groovy executor which takes an URL as argument and executes the content (Groovy script) of that URL in its classpath:
com.actelion.research.orbit.imageAnalysis.tasks.GroovyExecutor <URL>
We use this method to execute Groovy code within a Git repository.
Here is another example which shows how to iterate over all annotations of a set of images and output the class ratios for all annotations of all images, thus the output has the form
Image,Annotation,classRatio1, …classRatioN
import com.actelion.research.orbit.beans.RawAnnotation
import com.actelion.research.orbit.beans.RawDataFile
import com.actelion.research.orbit.imageAnalysis.dal.DALConfig
import com.actelion.research.orbit.imageAnalysis.models.ImageAnnotation
import com.actelion.research.orbit.imageAnalysis.models.OrbitModel
import com.actelion.research.orbit.imageAnalysis.utils.ClassificationResult
import com.actelion.research.orbit.imageAnalysis.utils.OrbitHelper
import com.actelion.research.orbit.imageAnalysis.utils.OrbitLogAppender
import groovyx.gpars.GParsPool
/**
* Iterates over all annotations and outputs classification results per image x annotation.
*/
def model = OrbitModel.LoadFromOrbit(2249052) // model id (stored on server)
def searchStr = "lung MT" // replace search string
OrbitLogAppender.GUI_APPENDER = false // no GUI (error) popups
DALConfig.getImageProvider().authenticateUser("root", "omero"); // optional: otherwise the scaleout user defined in OrbitOmero.conf is used
images = OrbitHelper.searchImages(searchStr)
StringBuilder sb = new StringBuilder("Image\tAnnotation")
model.classShapes.each { // append header
sb.append("\t"+it.name)
}
sb.append("\n")
GParsPool.withPool {
images.eachParallel { // attention: images will be computed in parallel, thus the output order is shuffled
RawDataFile rdf = it
StringBuilder sbImage = new StringBuilder()
DALConfig.getImageProvider().LoadRawAnnotationsByRawDataFile(rdf.rawDataFileId, RawAnnotation.ANNOTATION_TYPE_IMAGE).each {
ImageAnnotation anno = new ImageAnnotation(it)
ClassificationResult res = OrbitHelper.Classify(rdf, model, null, 1, anno.getFirstShape())
String s = rdf.fileName + "\t" + anno.description
res.normalizeRatio().each {
s += "\t" + it.value
}
s += "\n"
sbImage.append(s)
print s
}
sb.append(sbImage.toString())
}
}
println "\n\n\nResult:\n"
println sb.toString()
